Pcr Primer Pair

Pcr Primer Design In Silico Pcr And Oligonucleotides
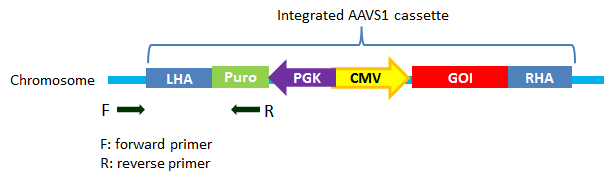
vs1 Int5 Pcr Primer Pairs Ge Origene

Untitled Document
Www Jstor Org Stable
101 Things You Maybe Didn T Know About Macvector 37 Testing Pcr Primer Pairs Using Primer3

Using Pcr Primers With Recombinase Polymerase Amplification
Note the two sets of Fabpi primers are used as internal controls.

Pcr primer pair. If only one primer is available, a. From the results of the PCR, TestPrime computes coverages for each taxonomic group in all of the taxonomies offered by SILVA. These primers are typically between 18 and 24 bases in length, and must code for only the specific upstream and downstream sites of the sequence being amplified.
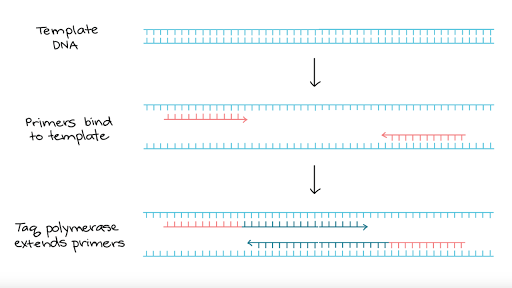
Primers are always specified 5' to 3', left to right. A primer is a short, single-stranded DNA sequence used in the polymerase chain reaction (PCR) technique. Inverse PCR • Inverse PCR (Ochman et al., 19) uses standard PCR (polymerase chain reaction)- primers oriented in the reverse direction of the usual orientation.
QSTAR qPCR primer pairs against Homo sapiens gene HPRT1. Real-time PCR primer assays consist of unlabeled PCR primer pairs for use with dye-based chemistry such as SYBR Green or EvaGreen. The direction of both forward and reverse primer should be 5′ to 3′.
The accuracy of these analyses depends strongly on the choice of primers. Ideally, primer Tm values should be near to the extension temperature. This length is long enough for adequate specificity and short enough for primers to bind easily to the template at the annealing temperature.
(I know that these need to be synthesised by the primer also for correct PCR technique) The boxed sets of nucleotides are the primer options I have at hand. The overall coverage and phylum spectrum of 175 primers and 512 primer pairs w …. Hence, both primers should be complementary to the sequences that flank the DNA fragment.
As its name implies, a PD consists of two primer molecules that have attached (hybridized) to each other because of strings of complementary bases in the primers. In this lecture, I explain how to design working primers for use in PCR. Primers should also be free of strong secondary structures and self-complementarity.
The basic guidelines for the successful design of PCR primers are described below. Primers for PCR and sequencing should have a GC content between 40 and 60%, with the 3′ of a primer ending in C or G to promote. Another important feature of a qPCR primer pair is the length of the primers.
Ta can be optimized by doing a temperature gradient PCR, starting at 5°C below the lowest Tm of the primer pair;. Too small a qPCR primer (<18bp) can increase the likelihood that it will bind to elsewhere in the genome, ie it is not specific enough to the target of interest. Primer blast works only a specificity check when a target template and both primers are given.
1 vial of lyophilized qSTAR qPCR primer mix (1 nmol each primer, sufficient for 0 reactions) 5S Human qPCR Primer Pair MP. The PCR primer desgin tool analyses the entered DNA sequence and chooses the optimum PCR primer pairs. Ideally, each primer should have a length of 18–22bps;.
The primer and Mg 2+ concentration in the PCR buffer and annealing temperature of the reaction may need to be optimized for each primer pair for efficient PCR. Custom primers The accuracy of design and synthesis of a primer pair is the most important consideration to generate good PCR performance data. Size depends on the sequence from which u designed primers.
So I want to choose the correct set of pair of primers to amplify the ORF of the gene that corresponds to amino acids in a protein. Therefore, it is possible to add sequence elements, like restriction sites, to the 5'-end of the primer molecule. QSTAR human and mouse U6 control miRNA primer pairs--0 reactions.
Poor design choices, erroneous or truncated sequences, and ineffective purification can lead to unusable results. I mean, to amplify my cDNA I should make a reaction mix with for example Taq DNA polymerase and only vary the concentrations of the primer pair and then afterwards verify optimal concentration on an agarose gel. Primers for PCR and sequencing should be between 18 to 25 nucleotides in length.
However, the most important considerations for primer design should be their T m value and specificity. If necessary, use a temperature gradient to further optimize and empirically determine the ideal annealing temperature for each template-primer pair combination. List of primers and probes labeled for EUA use and distributed by the International Reagent Resource may be used for viral testing with the CDC 19-nCoV Real-Time RT-PCR Diagnostic Panel.
PrimePCR™ PCR Primers, Assays, and Arrays. Primer Pair Matching Primers work in pairs – forward primer and reverse primer. Another issue with the 3’ primer ends is the possibility of homologies within the primer pair, leading to the dreaded primer-dimer effect.
For ex, u have sequence of 800 bp of DNA and u designed primer from starting and ending sequence of thatDNA then the PCR product size. 16S ribosomal RNA gene (rDNA) amplicon analysis remains the standard approach for the cultivation-independent investigation of microbial diversity. For single primers (determination of primer Tm) you can choose the Tm calculator for PCR.
Resources and interim guidelines for laboratory professionals working with specimens from persons with coronavirus disease 19 (COVID-19). DOWNLOAD Oligo Explorer 1.1.2 for Windows. DNA polymerase starts synthesizing new DNA from the 3’ end of the primer.
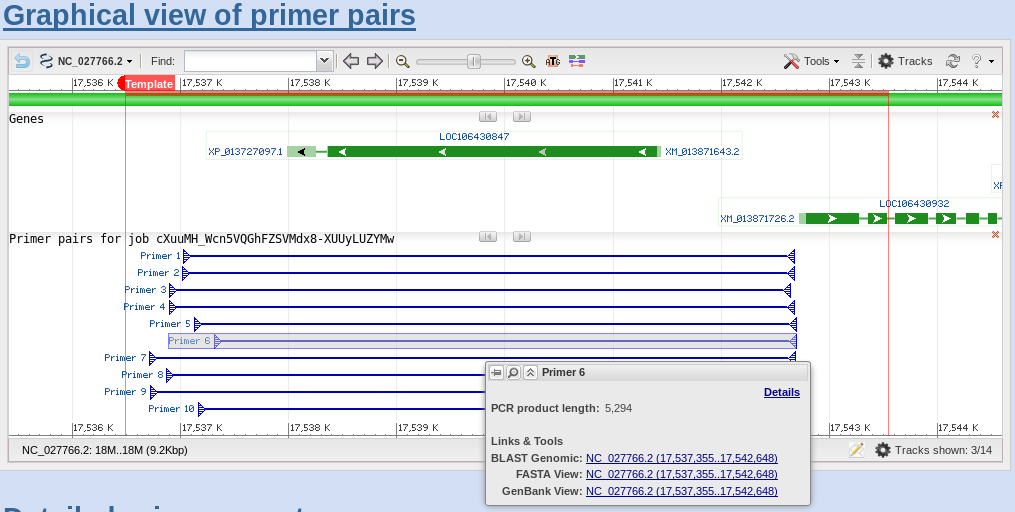
Run a PCR using the synthesized primer pairs. Gene-specific synthetic DNA template designed to give a positive real-time PCR result when used with the corresponding primer assay. Enable search for primer pairs specific to the intended PCR template ? With this option on, the program will search the primers against the selected database and determine whether a primer pair can generate a PCR product on any targets in the database based on their matches to the targets and their orientations.
DNA synthesis is the production of short, single-stranded DNA molecules (called primers or oligonucleotides) often used in the polymerase chain reaction (PCR) and DNA amplification for Sanger sequencing applications. The annealing temperature gradient should start with temperature 6-10°C lower than annealing temperature generated by the calculator and increased up to the extension temperature (two-step PCR). Each primer is shown in the location it would anneal to its template strand.
The primers are meticulously designed using OriGene's proprietary primer design algorithm developed from > 10,000 qPCR experiments. To do the PCR for the primer concentrations, it’s just a regular PCR right?. A primer dimer (PD) is a potential by-product in the polymerase chain reaction (PCR), a common biotechnological method.
Primer pairs are available for measuring the expression of specific human, mouse and rat mRNAs by RT-PCR. Multiplex PCR • Multiplex PCR is a variant of PCR which enabling simultaneous amplification of many targets of interest in one reaction by using more than one pair of primers. If you will be including a restriction site at the 5’ end of your primer, note that a 3-6 base pair "clamp" should be added upstream in order for the enzyme to cleave efficiently (e.g.
This analyzer requires at least 2 primer sequences in the input field. IDT recommends that you aim for PCR primers between 18 and 30 bases;. This often leads to primer-dimer formation.
Choosing suitable primers is an early crucial step in your quantitative reverse transcriptase PCR experiment - QRT-PCR. For amplification and detection of a well conserved region of the mitochondrial cytochrome c oxidase III (cox3) gene for Plasmodium species with a FAM labeled probe, forward primer, and reverse primer. Adjust individual primer pair concentrations in the multiplex pool if necessary, to balance the coverage of all primer pairs.
Probe assays for real-time PCR and Droplet Digital PCR include PCR primers and a dual-labeled fluorescent probe with your choice of fluorophore. At lower temperatures, PCR primers can anneal to each other via regions of complementarity, and the DNA polymerase can extend the annealed primers to produce primer dimer, which often appears as a diffuse band of approximately 50–100bp on an ethidium bromide-stained gel. Two sites offer software is based on the Primer3 program for design overlapping PCR primer pair sets - Multiple Primer Design with Primer 3 and Overlapping Primersets PHUSER (Primer Help for USER) - Uracil-Specific Exision Reagent (USER) fusion is a recently developed technique that allows for assembly of multiple DNA.
Two primers must be designed for PCR;. Otherwise primer selection from scratch is similar to that for a standard qualitative PCR experiment with some small variations. Usually bp is a common choice.
In the p rimer Pair s pecificity checking Parameters section, select the appropriate source organism. A total of 306,800 primers covering most known human and mouse genes can be accessed from the PrimerBank database, together with information on these primers such as T(m), location on the transcript and amplicon size. If the primers do not pass the test, synthesize the backup pair and repeat the check step until a suitable pair is found.
The forward primer and the reverse primer. Verify that your primers are designed and ordered in the correct orientation. QSTAR qPCR Primer Pairs are designed for SYBR Green-based real-time qPCR.
Primer pairs should be checked for complementarity at the 3'-end. Bases at the 5'-end of the primer are less critical for primer annealing. Reusing a tested primer pair from a repository or publication can save you some time.
The start and stop codons are underlined. The primer design algorithm has been extensively tested by real-time PCR experiments for PCR specificity and efficiency. Primer-dimer is when the PCR product obtained is the result of amplification of the primers themselves.
In selecting appropriate primers, a variety of constraints on the primer and amplified product sequences are already considered and taken as default values. The Fabpi primers amplify sequences from the endogenous mouse intestinal fatty acid binding protein gene. PCR primer design.
When individual primer pairs are multiplexed in a single PCR reaction, it is important to evaluate the breadth and depth of coverage of each primer pair for uniformity. The analyzer accepts text and table format (can be copied from an Excel file, for example). In addition, PCR efficiency can be improved by additives that promote DNA polymerase stability and processivity or increase hybridization stringency, and by using strategies that reduce.
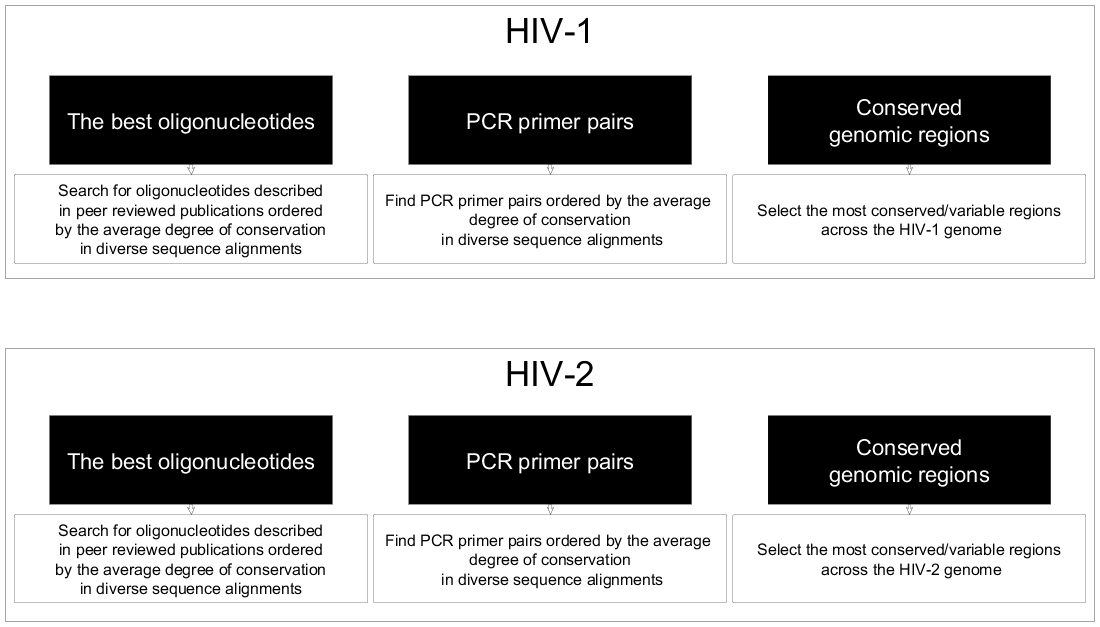
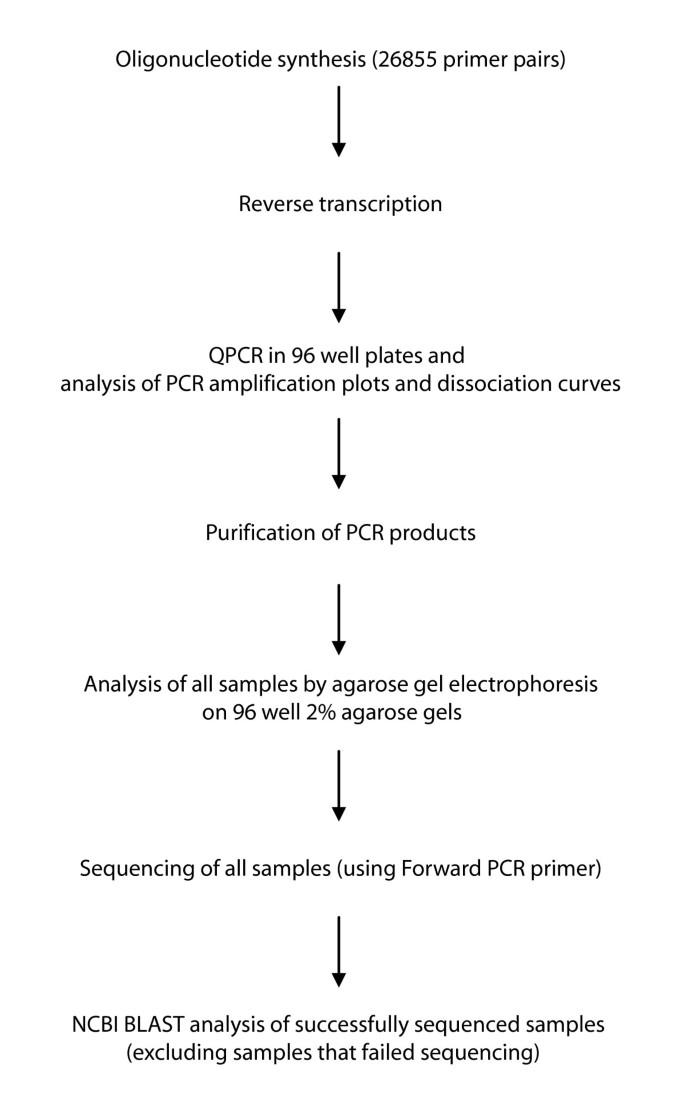
Primers are synthetic DNA strands of about 18 to 25 nucleotides complementary to 3’ end of the template strand. Which primer pair would best amplify the target region?. We have tested 26,855 primer pairs that correspond to 27,681 mouse genes by Real Time PCR followed by agarose gel electrophoresis and sequencing of the PCR products.
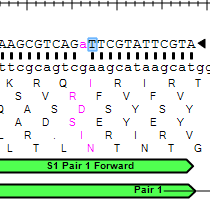
Four different pairs of PCR primers (in blue) are shown below. For each gene, at least one primer pair has been designed and in many cases alternative primer pairs exist. Design your PCR primers to conform to the following guidelines:.
These coverages can then be inspected in our taxonomy browser, making it easy to quickly identify strengths and weaknesses of a particular pair of primers. If you are unfamiliar with PCR, watch the following video:. To process some of your data.
If Tm values are calculated to be greater than the extension temperature, a two-step PCR program (combining annealing and extension into one step) can be employed;. Write or paste your primer sequences to the input field (upper window). Furthermore, we show that the ITS86F/ITS4 primer pair outperforms other primer pairs tested in terms of in silico primer efficiency, PCR efficiency, coverage, number of reads and number of species-level operational taxonomic units (OTUs) obtained.
The polymerase chain reaction (PCR) uses a pair of custom primers to direct DNA elongation toward each-other at opposite ends of the sequence being amplified. The maximum difference allowed is 3 C. Since they are used in the same PCR reaction, it shall be ensured that the PCR condition is suitable for both of them.
It is generally accepted that the optimal length of PCR primers is 18-22 bp. Test its efficiency and specificity by analyzing an gel electrophoresis result or a high resolution melting analysis (HRMA). Enter one or both primer sequences in the Primer Parameters section of the form.
Our primer collection covers the entire human and mouse genomes. This enables Disqus, Inc. These traits push the ITS86F/ITS4 primer pair forward as highly suitable for studying fungal.
In the Primer Pair Specificity Checking Parameters section, select the appropriate source Organism and the. You either can use the default constraint values or modify those values to customise. ONE OR MORE PRIMER SEQUENCES Go to the Primer BLAST submission form.
Primer Pairs are now available for several areas of research and have been utilized in qualitative RT-PCR analyses 1-10 (Figure 1. One critical feature is their annealing temperatures, which shall be compatible with each other. PCR Genotyping Primer Pairs The following primer pairs will amplify sequences present as a single copy in the mouse genome with the Universal Genotyping Protocol.
R&D Systems' PCR Primer Pairs are carefully designed and tested, so you spend less time optimizing PCR. The region of DNA amplified is determined by an exact match of the primer to its complimentary bases on a given DNA strand. Primer pairs should have a Tm within 5°C of each other Primer pairs should not have complementary regions Note:.
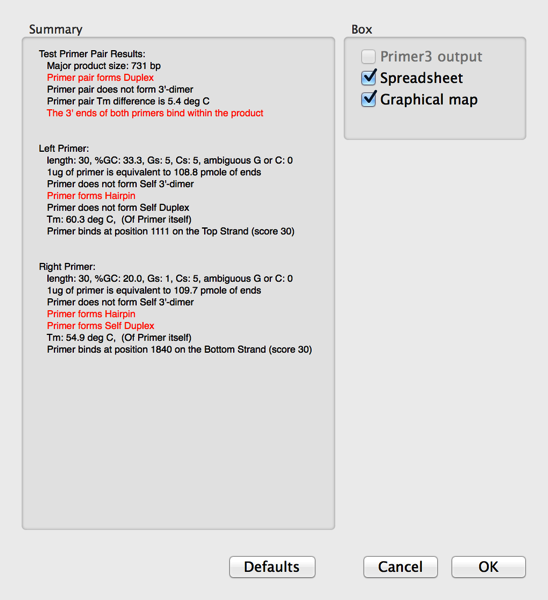
Testing Pairs Of Pcr Primer
Nested Pcr
Welcome To Hivoligodb
A Comprehensive Collection Of Experimentally Validated Primers For Polymerase Chain Reaction Quantitation Of Murine Transcript Abundance Bmc Genomics Full Text
Bmcgenomics Biomedcentral Com Track Pdf 10 1186 1471 2164 9 633
Pcr Primer Design Dnastar
How To Design Primers For Pcr
Q Tbn 3aand9gcqksw6z5yfg9k6y7nfzbgoh65zaj7edursq Hmswh5zid18pniz Usqp Cau
Pcr Primer Design Dnastar

Development Of Genome Wide Insertion And Deletion Polymorphism Markers From Next Generation Sequencing Data In Rice Rice Full Text

Pcr Primer Pair And Their Sequences Download Table

Multiplexed Identification Of Blood Borne Bacterial Pathogens By Use Of A Novel 16s Rrna Gene Pcr Ligase Detection Reaction Capillary Electrophoresis Assay Journal Of Clinical Microbiology
A Plastome Primer Set For Comprehensive Quantitative Real Time Rt Pcr Analysis Of Zea Mays A Starter Primer Set For Other Poaceae Species Springerlink

Rt Pcr Primer Pairs And Pcr Product Sizes Download Table
Www Jove Com Pdf 590 Jove Protocol 590 Detection Retrotransposition Activity Hot Line 1s Long Distance
Birch Designing Pcr Primers To Amplify A Gene From Genomic Dna

Literature Based Manually Curated Database Of Pcr Primers For The Detection Of Antibiotic Resistance Genes In Various Environments Sciencedirect
Polymerase Chain Reaction Pcr Article Khan Academy

Pcr Based Assay For Mating Type And Diploidy In Chlamydomonas Biotechniques
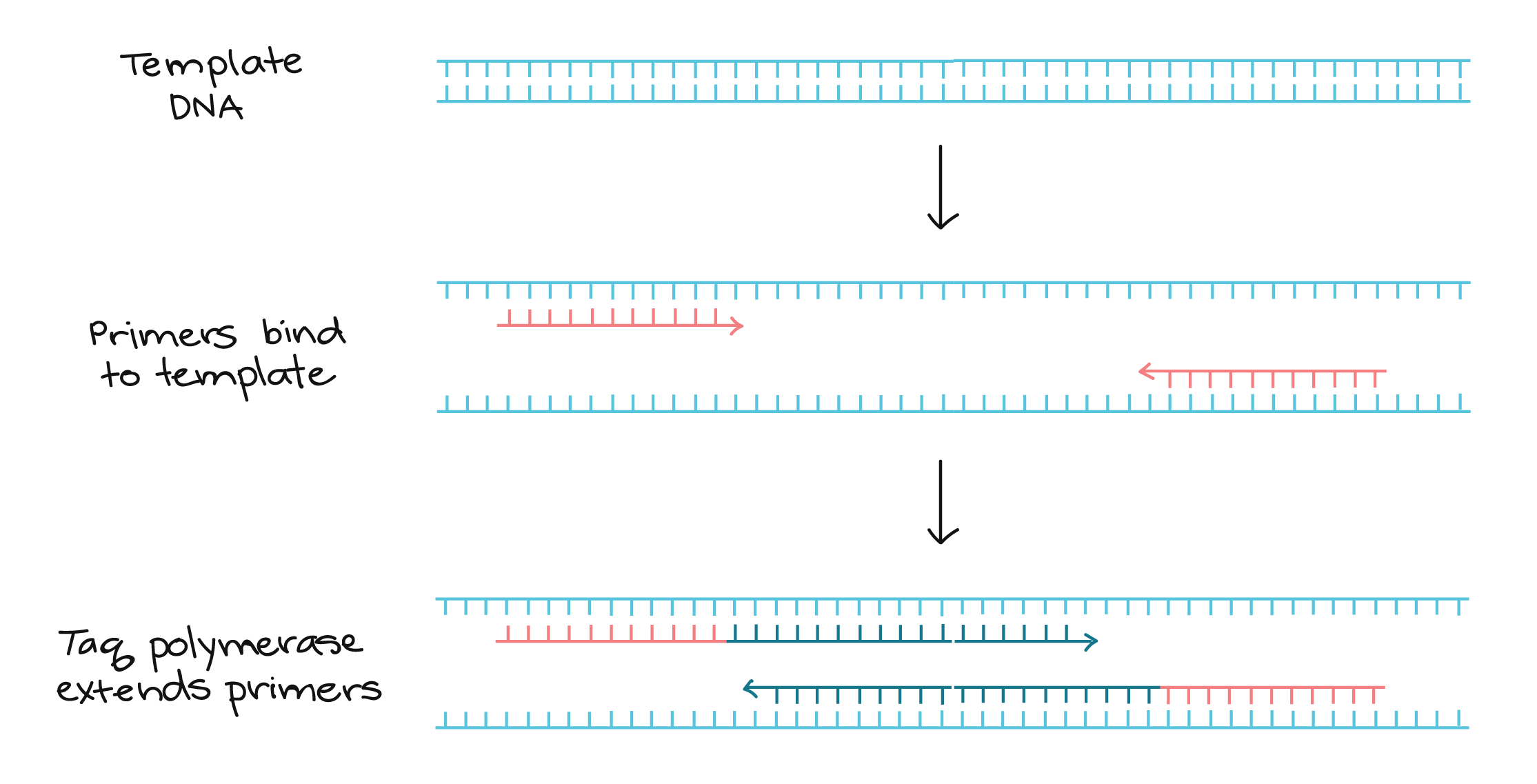
Polymerase Chain Reaction Pcr Article Khan Academy
Plos One Pcr Artifact In Testing For Homologous Recombination In Genomic Editing In Zebrafish

How To Select Primers For Polymerase Chain Reaction

Rt Pcr Primer Pairs Download Table

Table 1 From Family Specific Vs Universal Pcr Primers For The Study Of Mitochondrial Dna In Plants Semantic Scholar

Which Pair Of Primers Should Be Used To Amplify The Orf In Pcr Biology Stack Exchange
In Silico Pcr Primer Design And Gene Amplification Benchling
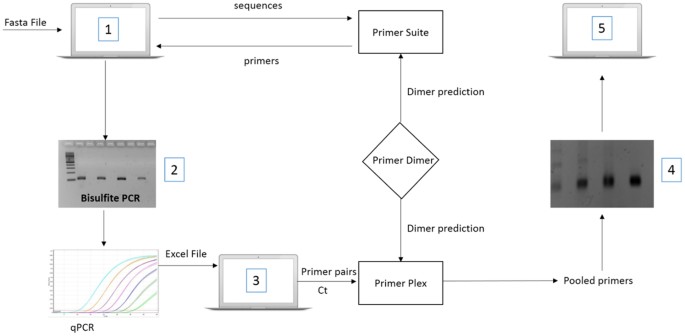
Primersuite A High Throughput Web Based Primer Design Program For Multiplex Bisulfite Pcr Scientific Reports
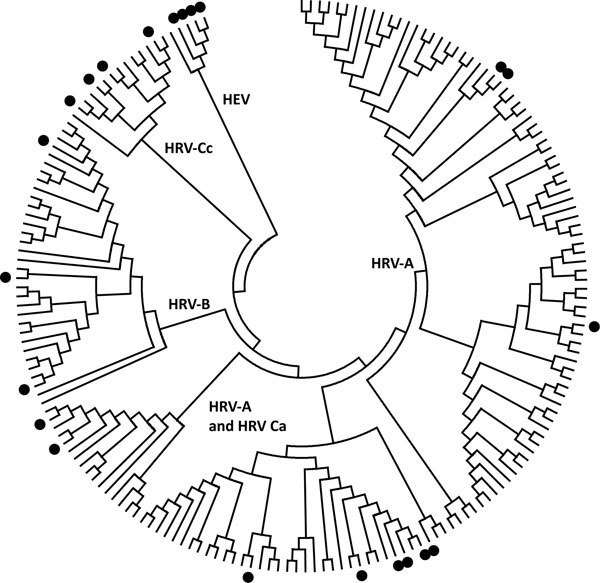
Figure Usefulness Of Published Pcr Primers In Detecting Human Rhinovirus Infection Volume 17 Number 2 February 11 Emerging Infectious Diseases Journal Cdc

How To Create Real Time Pcr Primers Using Primer Blast
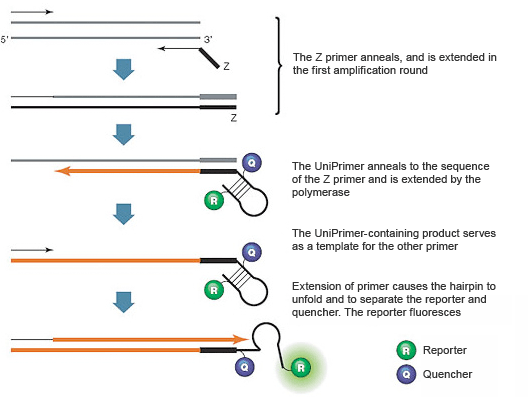
Introduction To Pcr Primer Probe Chemistries Lsr Bio Rad
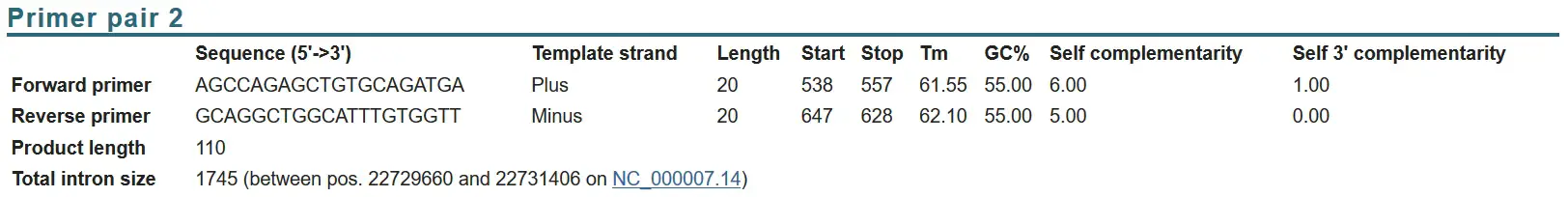
How To Create Real Time Pcr Primers Using Primer Blast
Q Tbn 3aand9gcqzninymvxcoxl6ityd4zga 5jbhu67honfuwvj3pcas6qidmae Usqp Cau

Pcr Primer Design

How To Select Primers For Polymerase Chain Reaction

Pcr Product Size And Pcr Primer Pairs Used To Amplify Ppo Genes Download Table

Tetra Primer Arms Pcr Optimization To Detect Single Nucleotide Polymorphism Of The Klf14 Gene

Bme105h1 Lecture Notes Winter 15 Lecture 8 Gc Content Complementary Dna Thought Experiment
Application Of Pcr Amplicon Sequencing Using A Single Primer Pair In Pcr Amplification To Assess Variations In Helicobacter Pylori Caga Epiya Tyrosine Phosphorylation Motifs Topic Of Research Paper In Veterinary Science
Q Tbn 3aand9gcrxpzzbwseiaace2zrsrmi21mtsxiztoadbzfj4cqv6110qknvm Usqp Cau

Polymerase Chain Reaction Pcr Article Khan Academy

Introduction To Pcr Primer Probe Chemistries Lsr Bio Rad
Digitalcommons Wustl Edu Cgi Viewcontent Cgi Article 1243 Context Open Access Pubs
2

American Journal Of Botany

Polymerase Chain Reaction Wikipedia
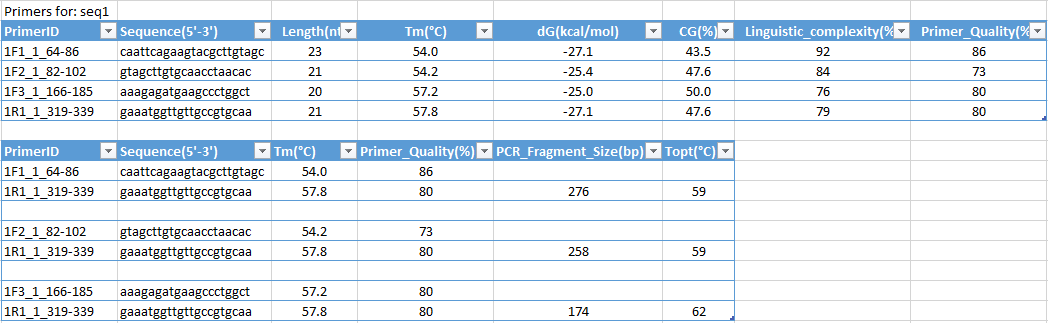
Pcr Primer Design In Silico Pcr And Oligonucleotides
Plos One Oligonucleotide Primers For Targeted Amplification Of Single Copy Nuclear Genes In Apocritan Hymenoptera
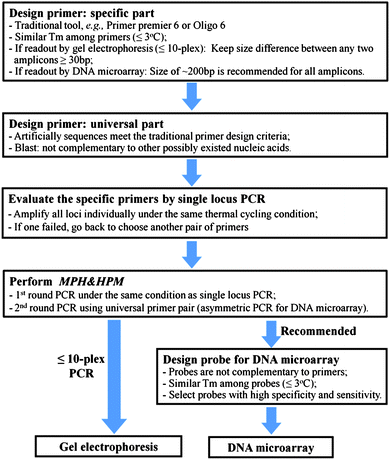
A Universal Multiplex Pcr Strategy For 100 Plex Amplification Using A Hydrophobically Patterned Microarray Lab On A Chip Rsc Publishing

Primer Blast A Tool To Design Target Specific Primers For Polymerase Chain Reaction Bmc Bioinformatics Full Text
Plos One Novel Pcr Primers For The Archaeal Phylum Thaumarchaeota Designed Based On The Comparative Analysis Of 16s Rrna Gene Sequences
Plos One A Novel Universal Primer Multiplex Pcr Method With Sequencing Gel Electrophoresis Analysis

Primer3 Bioz Ratings For Life Science Research
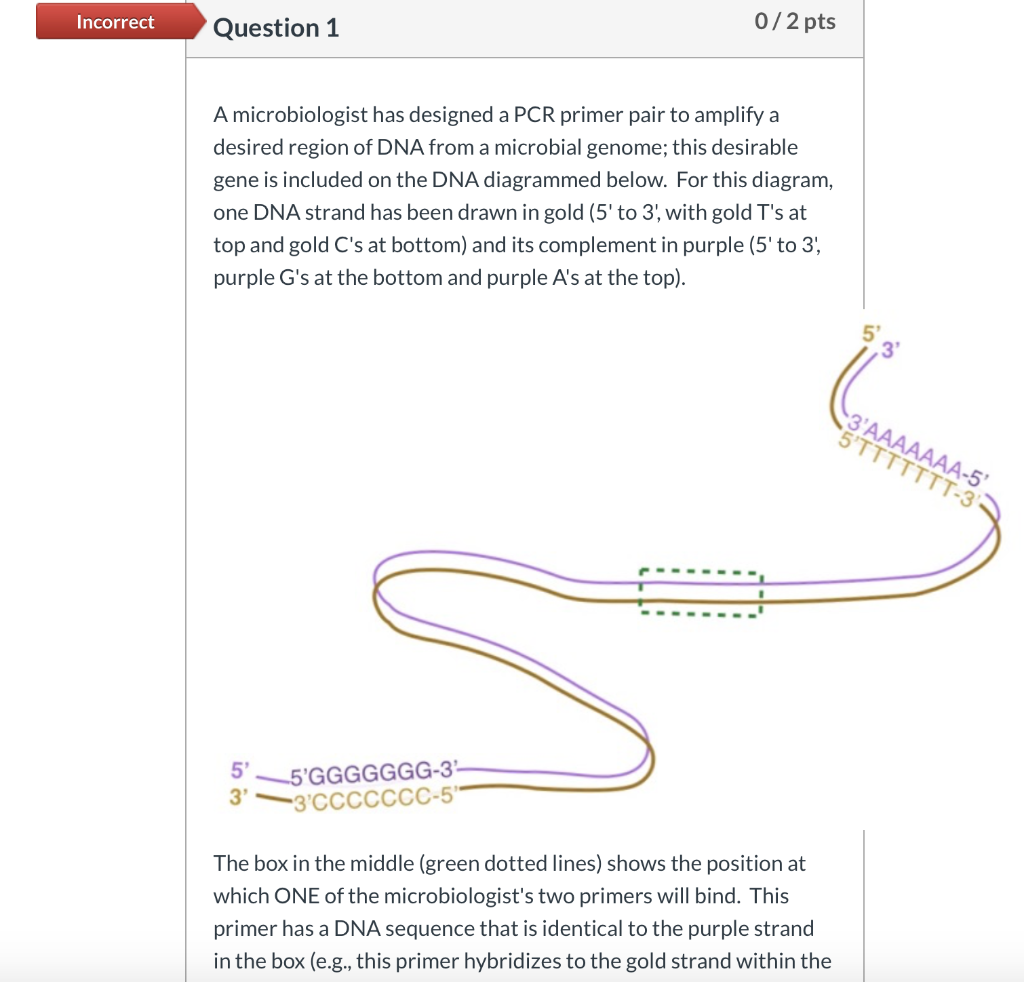
Solved Incorrect Question 1 0 2 Pts A Microbiologist Has Chegg Com

A Bioinformatics Workflow For The Evaluation Of Rt Qpcr Primer Specificity Application For The Assessment Of Gene Expression Data Reliability In Toxicological Studies Sciencedirect

Epa1 Pcr Primer Set For Bacterial Dna Amplification Kit For Detecting And Or Identifying Bacterial Species And Method For Detecting And Or Identifying Bacterial Species Google Patents
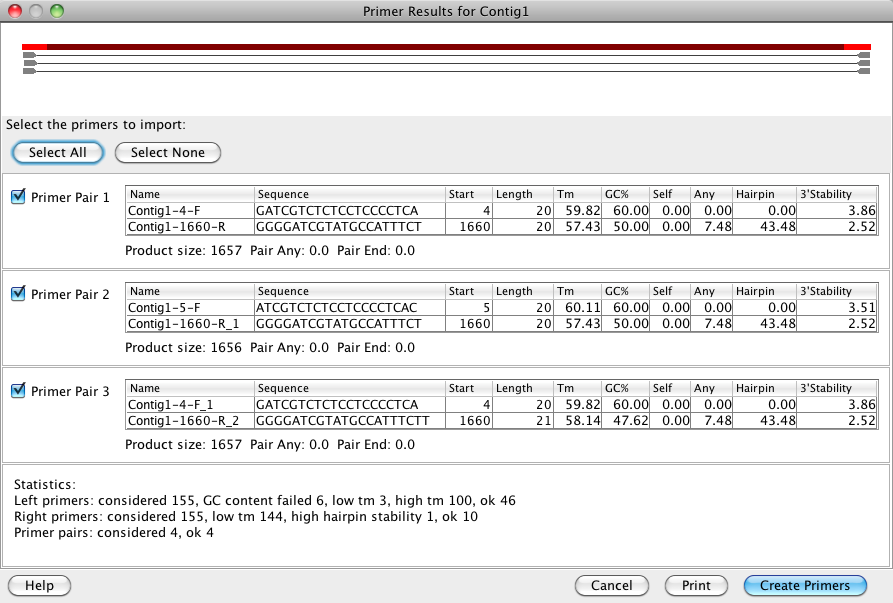
Primer Design In Codoncode Aligner

Table 1 Pcr Primer Pairs
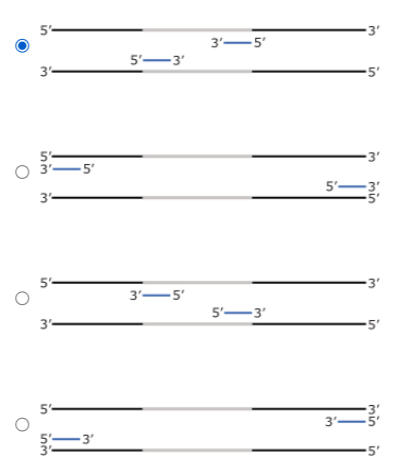
Answered Four Different Pairs Of Pcr Primers In Bartleby
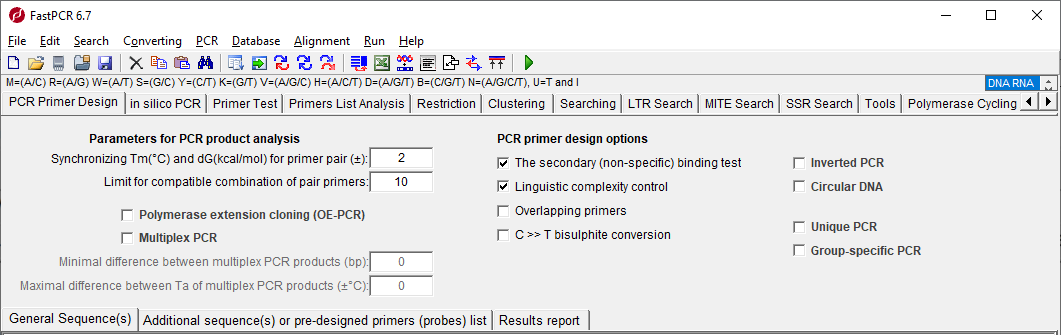
Fastpcr Manual

Introduction To Pcr Primer Probe Chemistries Lsr Bio Rad
Gtpb Github Io Elb19f Assets 04 Primer Design Practical Pdf

Pcr For Sanger Sequencing Thermo Fisher Scientific Es
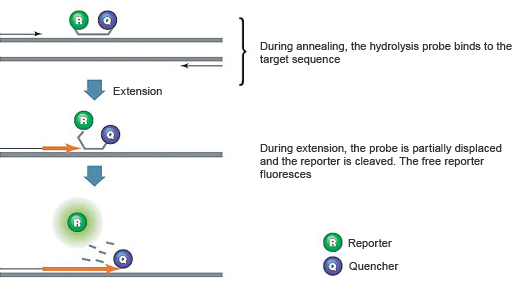
Introduction To Pcr Primer Probe Chemistries Lsr Bio Rad

Primer Pairs R D Systems
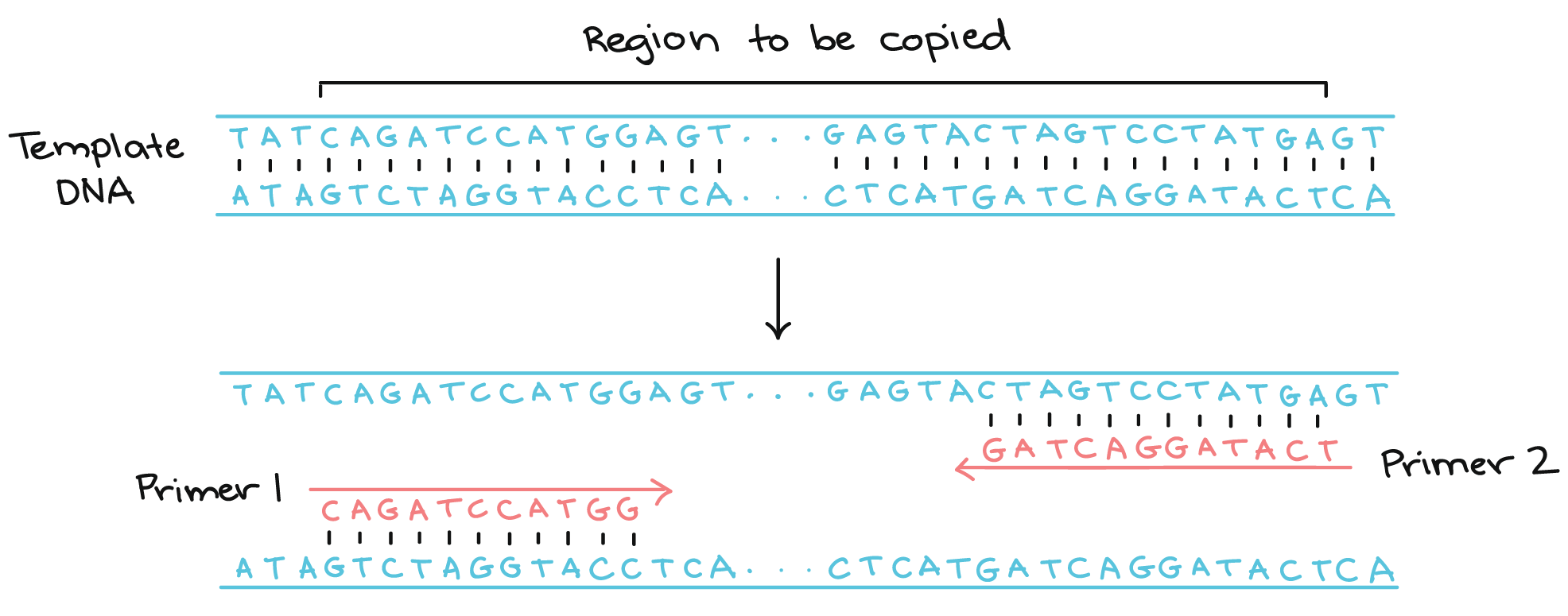
Polymerase Chain Reaction Pcr Article Khan Academy

Protocol Primer Design
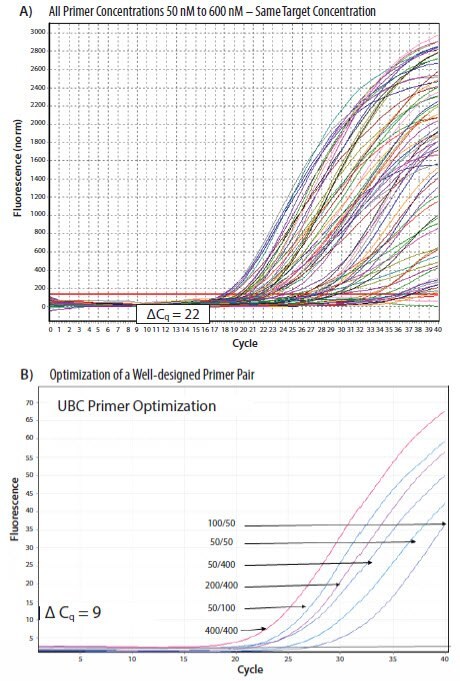
Primer Validation For Optimum Assay Performance Pcr Guide Sigma Aldrich

Primer Design Tutorial Geneious Prime
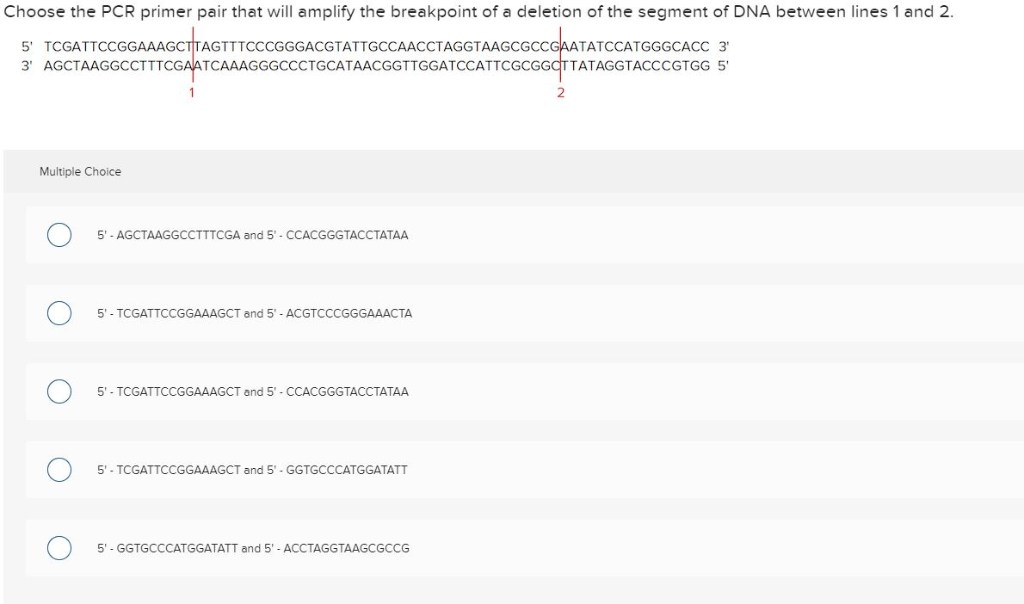
Solved Choose The Pcr Primer Pair That Will Amplify The B Chegg Com

Empirical Testing Of 16s Rrna Gene Pcr Primer Pairs Reveals Variance In Target Specificity And Efficacy Not Suggested By In Silico Analysis Applied And Environmental Microbiology
How To Design Primers For Pcr
Epublications Marquette Edu Cgi Viewcontent Cgi Article 10 Context Theses Open

Principles Of Direct Sequencing And Genomic Qpcr For Ge Open I

Pcr For Sanger Sequencing Thermo Fisher Scientific Es
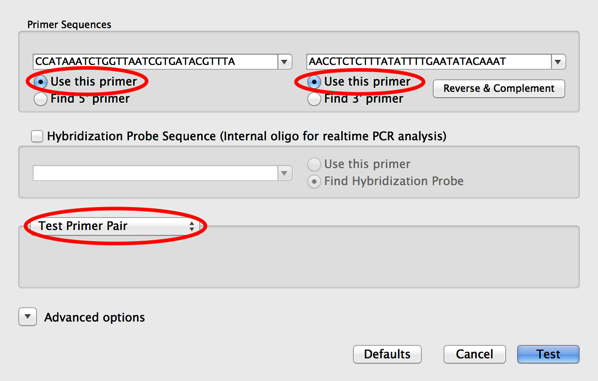
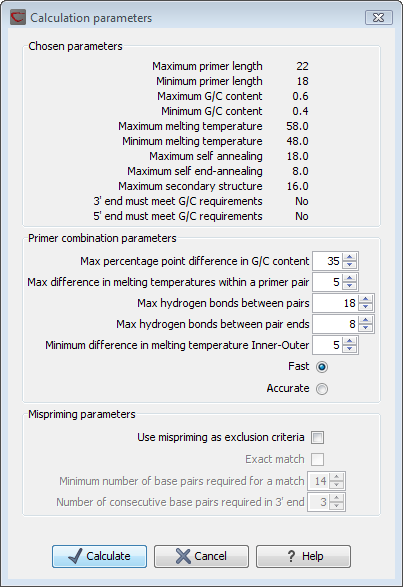
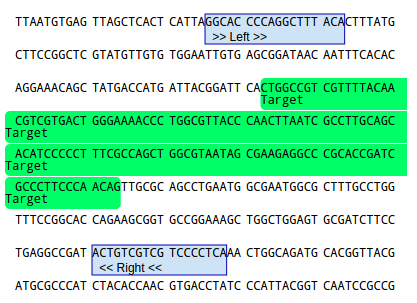
Test Pcr Primer Pairs Using The Primer Design Primer3 Interface

Untitled Document
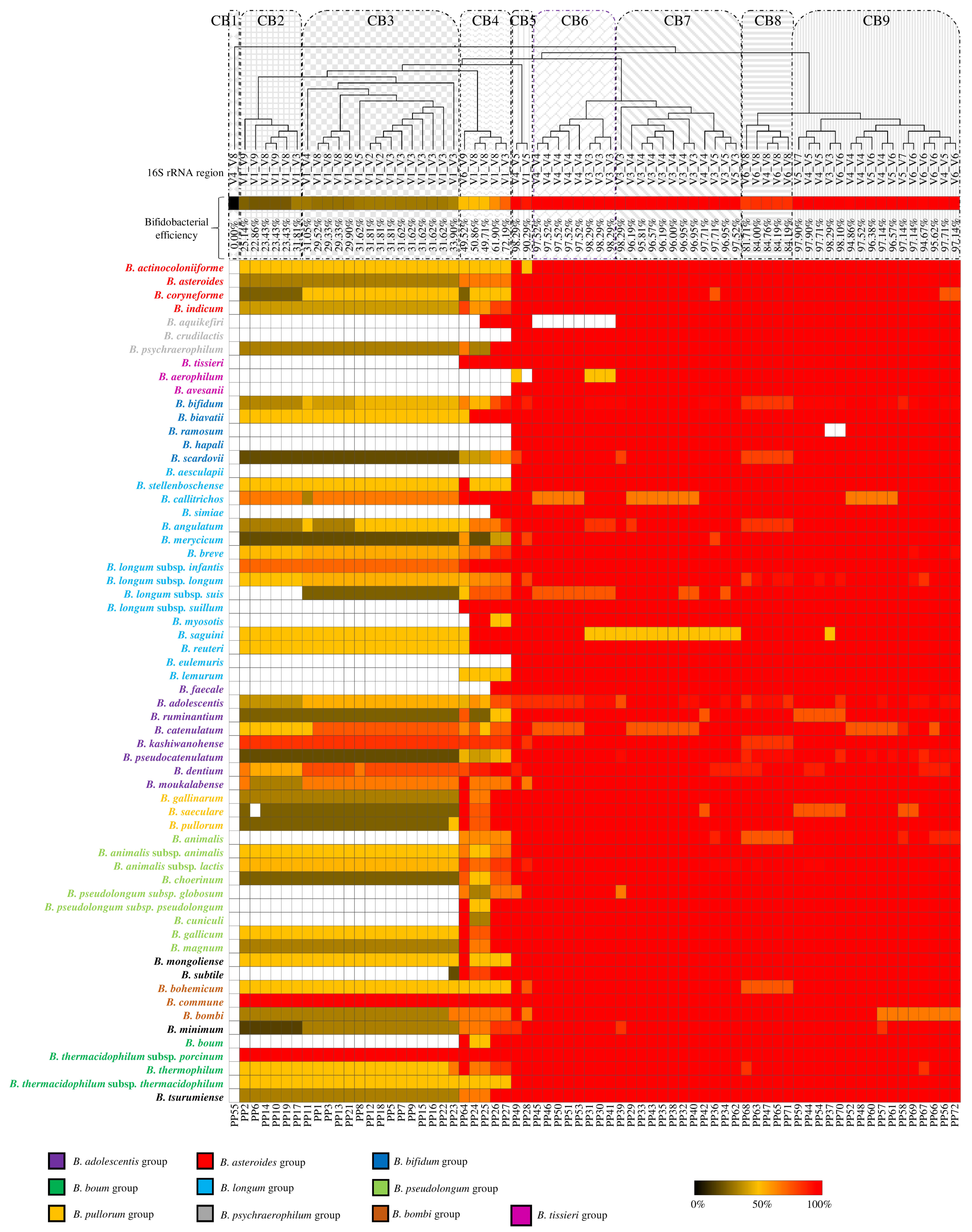
Microorganisms Free Full Text The Impact Of Primer Design On Amplicon Based Metagenomic Profiling Accuracy Detailed Insights Into Bifidobacterial Community Structure Html
2

Table S1 Sequences Of The Three Pcr Primer Pairs A B

Pcr Primer Pairs Used In This Study Download Table
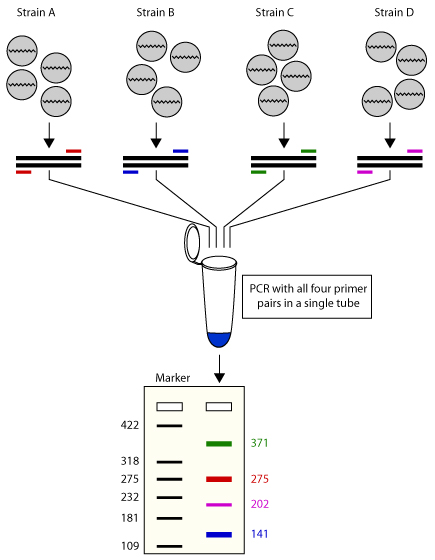
Multiplex Pcr An Overview Of Multiplex Pcr Assay Primer Design For Multiplexing Primer Design Software For Multiplex Pcr

Woa1 Pcr Primer Pair And Application Thereof Google Patents
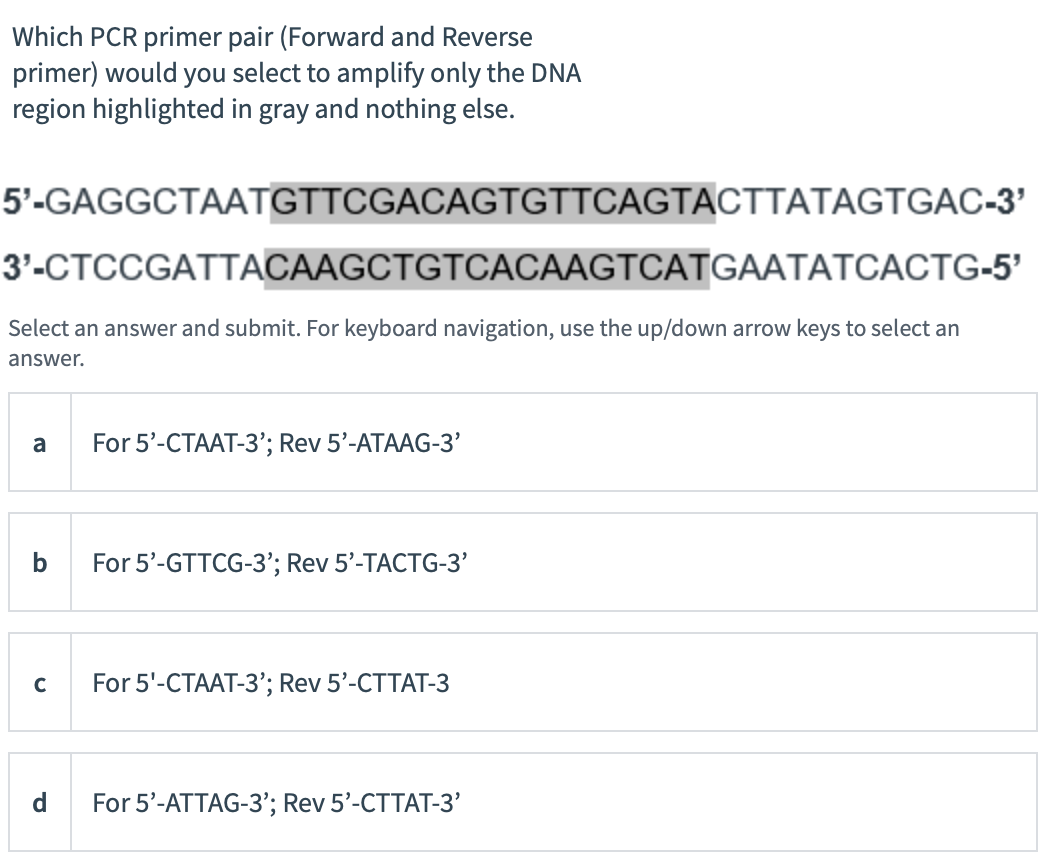
Solved Which Pcr Primer Pair Forward And Reverse Primer Chegg Com
Birch Designing Pcr Primers To Amplify A Gene From Genomic Dna
Q Tbn 3aand9gctvfxchhuftpc Zex Bfuwfrm9 C8 Dg3mp9u1pmqfbzgxvz Go Usqp Cau
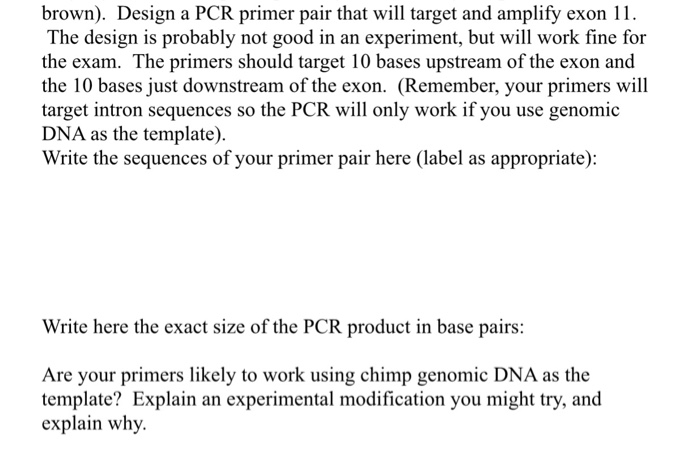
Solved Brown Design A Pcr Primer Pair That Will Target Chegg Com
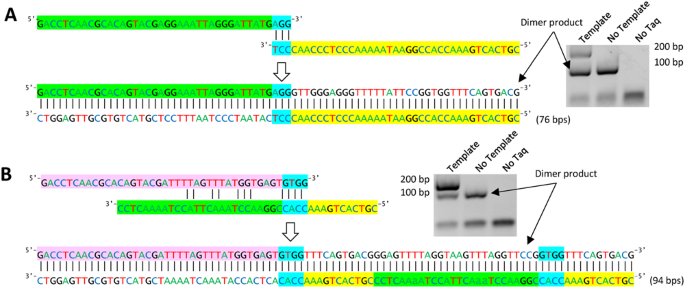
Primerroc Accurate Condition Independent Dimer Prediction Using Roc Analysis Scientific Reports

Multiplex Polymerase Chain Reaction An Overview Sciencedirect Topics
2

Primegensw3

Pcr From Setup To Cleanup A Beginner S Guide With Useful Tips And

Untitled Document
2

Linear After The Exponential Late Pcr An Advanced Method Of Asymmetric Pcr And Its Uses In Quantitative Real Time Analysis Pnas

Cpt Galaxy Training

Addgene Protocol How To Design Primers

Perlprimer

Bistro Primer Tool To Design And Validate Specific Pcr Primer Pairs For Phylogenetic Analysis Semantic Scholar



